Locus annotations
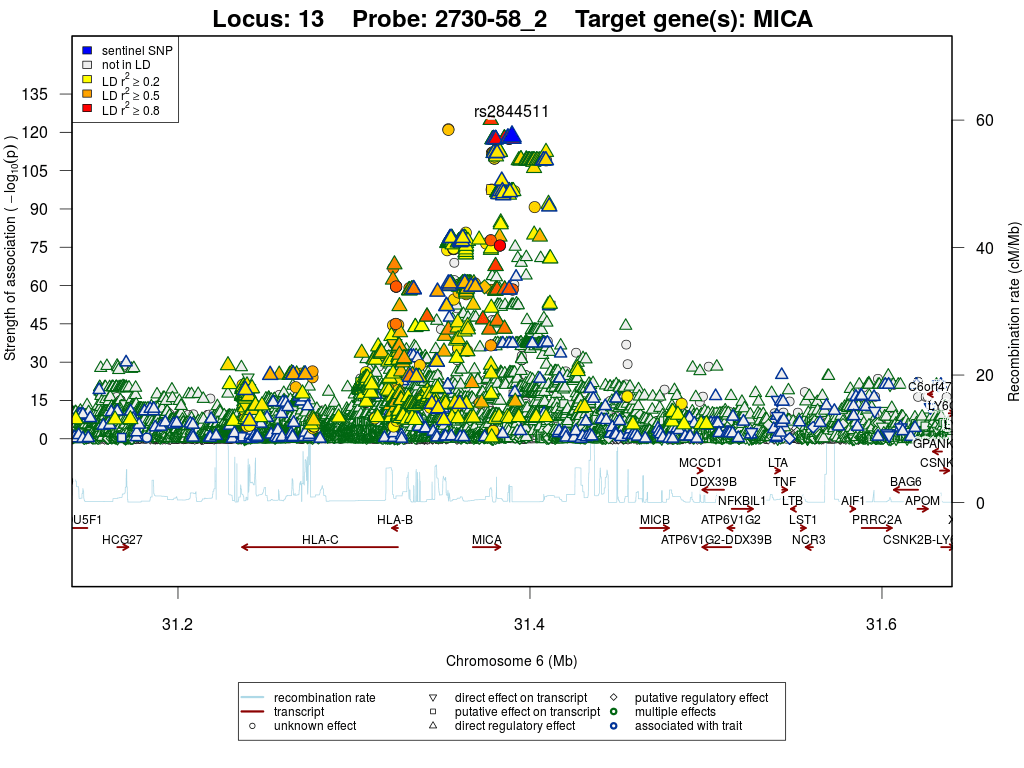
Locus 13
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MICA | cis | Discovery | rs2844511 | 6:31,389,784 | G/A | 0.40 | 995 | 0.922 | 0.034 | 3.8×10-119 | 1.860 | 2.8×10-109 | 7.7×10-51 |
| MICA | cis | Replication | rs2596530 | 6:31,387,373 | A/G | 0.16 | 337 | 0.616 | 0.089 | 2.7×10-11 | 1.370 | 7.9×10-10 | 1.7×10-4 |
Regional association plots
MHC class I polypeptide-related sequence A (MICA)
Boxplots and histograms for top associations
MHC class I polypeptide-related sequence A (MICA)
| Target (abbrv.) | MICA |
| Target (full name) | MHC class I polypeptide-related sequence A |
| Somalogic ID (Sequence ID) | SL005199 (2730-58_2) |
| Entrez Gene Symbol | MICA |
| UniProt ID | Q29983 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
| Pathway Studio |
|
All locus annotations are based on the sentinel SNP (rs2844511) and 9 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by | |
| Potentially regulated genes | |
| eQTL genes |
|
Results from other genome-wide association studies
| Trait | P | Study | Source |
|---|---|---|---|
| Gene expression | 2×10-62 | 20881960 (PMID) | GRASP2 nonQTL |
| cg23656755 (chr1:230203067) | 2.6×10-41 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg06606381 (chr12:133084921) | 1.2×10-39 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg01620082 (chr3:125678431) | 2×10-39 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg17862947 (chr12:133086950) | 6.5×10-38 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Height | 2.5×10-36 | 25282103 (PMID) | Supplemental file |
| Gene expression of MICA (probeID ILMN_1797929) in temporal cortex in Alzheimer's disease cases and controls | 2.2×10-27 | 22685416 (PMID) | GRASP2 eQTL |
| Gene expression of XRCC6 in blood | 4.2×10-24 | 21829388 (PMID) | GRASP2 eQTL |
| cg17169043 (chr6:170863821) | 4×10-20 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Gene expression of HCG27 in blood | 9×10-19 | 21829388 (PMID) | GRASP2 eQTL |
| Cervical cancer | 1.6×10-18 | 23482656 (PMID) | GRASP2 nonQTL |
| CERVICAL CANCER | 4×10-18 | 23482656 (PMID) | GWAS Catalog via SNiPA |
| Gene expression of MICA in normal prepouch ileum | 1.3×10-17 | 23474282 (PMID) | GRASP2 eQTL |
| Gene expression of TMEM154 in blood | 3.7×10-17 | 21829388 (PMID) | GRASP2 eQTL |
| Gene expression of MICA (probeID ILMN_1797929) in temporal cortex in non-Alzheimer's disease samples | 1.4×10-16 | 22685416 (PMID) | GRASP2 eQTL |
| cg02753903 (chr19:38943549) | 1.5×10-15 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Idiopathic membranous nephropathy | 1.9×10-14 | 21323541 (PMID) | GRASP2 nonQTL |
| cg22379212 (chr19:38944024) | 5.7×10-12 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Gene expression of MICA (probeID ILMN_1797929) in temporal cortex in Alzheimer's disease cases | 1.1×10-11 | 22685416 (PMID) | GRASP2 eQTL |
| Gene expression of LOC282956 in CEU-CHB-JPT-YRI lymphoblastoid cell lines | 1.2×10-11 | 17873874 (PMID) | GRASP2 eQTL |
| Triglycerides | 2.7×10-11 | 24097068 (PMID) | Supplemental file |
| cg01255021 (chr16:1094629) | 4.5×10-11 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Rheumatoid arthritis | 1.1×10-10 | 20453842 (PMID) | GRASP2 nonQTL |
| HIV-1 control (viral load at set point) | 1.1×10-10 | 20041166 (PMID) | GRASP2 nonQTL |
| Gene expression of LOC285835 [probe 230937_at] in lymphoblastoid cell lines | 1.1×10-10 | 17873877 (PMID) | GRASP2 eQTL |
| cg04788957 (chr10:105438032) | 1.3×10-10 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| HIV-1 control | 2.1×10-9 | 21051598 (PMID) | GRASP2 nonQTL |
| Lupus erythematosus, systemic | 3.6×10-9 | pha002848 (dbGaP) | dbGaP via SNiPA |
| cg10154826 (chr6:17601018) | 3.7×10-9 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg09408951 (chr1:187673755) | 6.2×10-9 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg25652859 (chr20:57427388) | 1.2×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Gene expression of LST1///NCR3 in blood | 1.2×10-8 | 21829388 (PMID) | GRASP2 eQTL |
| cg20954971 (chr21:44593666) | 1.3×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg27443779 (chr11:14664769) | 1.4×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg19313311 (chr7:74058055) | 1.4×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg19589727 (chr20:57427786) | 1.6×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Height (adults) | 3.2×10-8 | 21194676 (PMID) | GRASP2 nonQTL |
| Behcet syndrome | 4.3×10-8 | pha002888 (dbGaP) | dbGaP via SNiPA |
| cg14564778 (chr20:57427532) | 5×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg03059896 (chr1:27560026) | 8.9×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg17841099 (chr1:153940066) | 9×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg06065549 (chr20:57427419) | 1.3×10-7 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg00943909 (chr20:57427966) | 1.3×10-7 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Total cholesterol | 1.5×10-7 | 23063622 (PMID) | GRASP2 nonQTL |
| cg14153654 (chr1:8001003) | 2×10-7 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| cg05496363 (chr11:66083790) | 2.6×10-7 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| HIV-1 seropositive nonprogression | 4.7×10-7 | 19115949 (PMID) | GRASP2 nonQTL |
| HIV-1 infection (natural long-term nonprogression) | 1.2×10-6 | 22238471 (PMID) | GRASP2 nonQTL |
| Height (adult females) | 1.5×10-6 | 21194676 (PMID) | GRASP2 nonQTL |
| HDL cholesterol | 1.7×10-6 | 24097068 (PMID) | Supplemental file |
| Differential exon level expression of HLA-B [probe 2948952] in peripheral blood mononuclear cells | 2.4×10-6 | 19222302 (PMID) | GRASP2 eQTL |
| Hip circumfence, adjusted for BMI | 3.6×10-6 | 25673412 (PMID) | Supplemental file |
| cholesterol | 4.6×10-6 | 24816252 (PMID) | gwas.eu via SNiPA |
| HIV-1-specific cross-reactive neutralizing activity | 1.3×10-5 | 23372753 (PMID) | GRASP2 nonQTL |
| Differential exon level expression of MICB [probe 2902359] in brain cortex | 1.3×10-5 | 19222302 (PMID) | GRASP2 eQTL |
| LDL cholesterol | 2.1×10-5 | 24097068 (PMID) | Supplemental file |
| Hip circumfence | 3.7×10-5 | 25673412 (PMID) | Supplemental file |
| Gene expression of MICA in CD4+ lymphocytes | 5.8×10-5 | 20833654 (PMID) | GRASP2 eQTL |
| Schizophrenia | 1×10-4 | 19197363 (PMID) | GRASP2 nonQTL |
| Type 2 diabetes | 1.5×10-4 | 22885922 (PMID) | Supplemental file |
| Crohn's disease | 2×10-4 | Supplemental file |