Locus 306
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RUXF | trans | Discovery | rs4420638 | 19:45,422,946 | A/G | 0.17 | 994 | -0.435 | 0.056 | 3×10-14 | -1.040 | 2.5×10-4 | 0.314 |
| RUXF | trans | Replication | rs4420638 | 19:45,422,946 | A/G | 0.12 | 337 | -0.263 | 0.104 | 0.012 | -1.050 | 0.031 | 0.126 |
| Apo E2 | cis | Discovery | rs4420638 | 19:45,422,946 | A/G | 0.17 | 994 | 0.419 | 0.056 | 1.9×10-13 | 1.070 | 1.9×10-12 | 1.5×10-13 |
| Apo E2 | cis | Replication | rs4420638 | 19:45,422,946 | A/G | 0.12 | 337 | 0.270 | 0.107 | 0.012 | 1.050 | 0.029 | 0.014 |
Regional association plots
Small nuclear ribonucleoprotein F (RUXF)
Apolipoprotein E (isoform E2) (Apo E2)
Boxplots and histograms for top associations
Small nuclear ribonucleoprotein F (RUXF)
| Target (abbrv.) | RUXF |
| Target (full name) | Small nuclear ribonucleoprotein F |
| Somalogic ID (Sequence ID) | SL009790 (5494-52_3) |
| Entrez Gene Symbol | SNRPF |
| UniProt ID | P62306 |
| UniProt Comment |
|
| Wiki Pathways |
|
| Reactome |
|
Apolipoprotein E (isoform E2) (Apo E2)
| Target (abbrv.) | Apo E2 |
| Target (full name) | Apolipoprotein E (isoform E2) |
| Somalogic ID (Sequence ID) | SL000277 (5312-49_3) |
| Entrez Gene Symbol | APOE |
| UniProt ID | P02649 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
| Wiki Pathways |
|
| Reactome |
|
All locus annotations are based on the sentinel SNP (rs4420638) and 4 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by |
|
Results from other genome-wide association studies
| Trait | P | Study | Source |
|---|---|---|---|
| Alzheimer's disease | 0×100 | 24162737 (PMID) | Supplemental file |
| Late onset Alzheimer's disease | 1×10-300 | 21460841 (PMID) | GRASP2 nonQTL |
| LDL cholesterol | 1.5×10-178 | 24097068 (PMID) | Supplemental file |
| Cholesterol, total | 1×10-149 | 24097068 (PMID) | GWAS Catalog via SNiPA |
| Triglycerides | 1.1×10-149 | 24097068 (PMID) | Supplemental file |
| C-reactive protein (CRP) | 8.8×10-139 | 21300955 (PMID) | GRASP2 nonQTL |
| C-reactive protein levels | 9×10-139 | 21300955 (PMID) | GWAS Catalog via SNiPA |
| Total cholesterol | 2.7×10-116 | 20686565 (PMID) | GRASP2 nonQTL |
| Total cholesterol (females) | 1×10-72 | 20686565 (PMID) | GRASP2 nonQTL |
| Metabolic syndrome domains (Multivariate analysis) | 1.7×10-57 | 22022282 (PMID) | GRASP2 nonQTL |
| Alzheimer's disease (late onset) | 1×10-39 | 17474819 (PMID) | GWAS Catalog via SNiPA |
| Sporadic late onset Alzheimer's disease | 1.1×10-39 | 17474819 (PMID) | GRASP2 nonQTL |
| Longevity (90 years and older) | 3×10-36 | 24688116 (PMID) | GWAS Catalog via SNiPA |
| APOB (apolipoprotein B) | 3.1×10-32 | 23100282 (PMID) | GRASP2 nonQTL |
| Metabolic syndrome domains (Atherogenic Dyslipidemia - PC1) | 1×10-31 | 22022282 (PMID) | GRASP2 nonQTL |
| APOE (apolipoprotein E) | 8.1×10-31 | 20442857 (PMID) | GRASP2 nonQTL |
| Lp-PLA2 activity | 4.9×10-30 | 22003152 (PMID) | GRASP2 nonQTL |
| Lipoprotein-associated phospholipase A2 activity and mass | 5×10-30 | 22003152 (PMID) | GWAS Catalog via SNiPA |
| Rate of age-related cognitive decline | 3.7×10-27 | 22054870 (PMID) | GRASP2 nonQTL |
| Cognitive decline | 4×10-27 | 22054870 (PMID) | GWAS Catalog via SNiPA |
| C-reactive protein | 5×10-27 | 19567438 (PMID) | GWAS Catalog via SNiPA |
| Longevity (85 years and older) | 2×10-26 | 24688116 (PMID) | GWAS Catalog via SNiPA |
| HDL cholesterol | 2.3×10-22 | 20686565 (PMID) | GRASP2 nonQTL |
| Lipoprotein-associated phospholipase A2 activity (Lp-PLA2) | 9.2×10-21 | 23118302 (PMID) | GRASP2 nonQTL |
| LDL cholesterol in serum | 1.2×10-20 | 18262040 (PMID) | GRASP2 nonQTL |
| Advanced age-related macular degeneration | 2×10-20 | 23455636 (PMID) | GRASP2 nonQTL |
| Age-related macular degeneration | 2×10-20 | 23455636 (PMID) | GWAS Catalog via SNiPA |
| Total cholesterol (males) | 6×10-20 | 20686565 (PMID) | GRASP2 nonQTL |
| Longevity | 2×10-16 | 21740922 (PMID) | GWAS Catalog via SNiPA |
| Lipid traits | 1×10-14 | 24023261 (PMID) | GWAS Catalog via SNiPA |
| Alzheimer's disease (age of onset) | 4.1×10-14 | 22005931 (PMID) | GRASP2 nonQTL |
| LDL cholesterol response to statins | 3.7×10-12 | 22368281 (PMID) | GRASP2 nonQTL |
| Alzheimer's disease biomarkers | 4×10-12 | 23419831 (PMID) | GWAS Catalog via SNiPA |
| Metabolic syndrome domains (Vascular Inflammation - PC1) | 5×10-12 | 22022282 (PMID) | GRASP2 nonQTL |
| High-sensitivity C-reactive protein (CRP) | 7.1×10-11 | 22291609 (PMID) | GRASP2 nonQTL |
| LDL cholesterol response to statins (first post-treatment LDL cholesterol) | 1.2×10-9 | 22368281 (PMID) | GRASP2 nonQTL |
| Advanced age-related macular degeneration (geographic atrophy) | 1.9×10-9 | 23455636 (PMID) | GRASP2 nonQTL |
| Advanced age-related macular degeneration (choroidal neovascularization) vs. no AMD | 2.8×10-8 | 23455636 (PMID) | GRASP2 nonQTL |
| Serum C-reactive protein | 2.9×10-7 | 21196492 (PMID) | GRASP2 nonQTL |
| Type 2 diabetes | 3.2×10-7 | 22885922 (PMID) | Supplemental file |
| APOB (apolipoprotein B) response after 40mg daily simvastatin treatment | 5.7×10-7 | 23100282 (PMID) | GRASP2 nonQTL |
| LDL cholesterol change with statins | 6.3×10-7 | 20339536 (PMID) | GRASP2 nonQTL |
| Metabolic syndrome domains (Central obesity) | 1.2×10-6 | 22022282 (PMID) | GRASP2 nonQTL |
| Coronary artery disease (CAD) | 1.5×10-6 | 20864672 (PMID) | GRASP2 nonQTL |
| Quantitative traits | 2×10-6 | 19197348 (PMID) | GWAS Catalog via SNiPA |
| Gene expression of CD81 in peripheral blood monocytes | 3.4×10-6 | 20502693 (PMID) | GRASP2 eQTL |
| Gene expression of MAD1L1 in blood | 3.5×10-6 | 21829388 (PMID) | GRASP2 eQTL |
| Waist circumference | 3.6×10-6 | 19557197 (PMID) | GRASP2 nonQTL |
| Waist-to-hip ratio, adjusted for BMI | 4.2×10-6 | 25673412 (PMID) | Supplemental file |
| LDL cholesterol response after 40mg daily simvastatin treatment | 6.4×10-6 | 23100282 (PMID) | GRASP2 nonQTL |
| Waist-to-hip ratio | 7.6×10-6 | 25673412 (PMID) | Supplemental file |
| Gene expression of CCDC86///GPR44 in blood | 9.1×10-6 | 21829388 (PMID) | GRASP2 eQTL |
| Total cholesterol change with statins | 1.6×10-5 | 20339536 (PMID) | GRASP2 nonQTL |
| cg23184690 (chr19:45429795) | 4.4×10-5 | 10.1101/033084 (DOI) | BIOS QTL cis-meQTLs |
| Coronary artery disease (CAD), combined control dataset | 1.7×10-4 | 17554300 (PMID) | GRASP2 nonQTL |
| Coronary heart disease (CHD) or coronary artery disease (CAD) | 2.1×10-4 | 22003152 (PMID) | GRASP2 nonQTL |
| Triglycerides (ln) | 3×10-4 | 23063622 (PMID) | GRASP2 nonQTL |
| Fasting glucose | 3.1×10-4 | 22885924 (PMID) | Supplemental file |
| Lp-PLA2 mass | 4.3×10-4 | 22003152 (PMID) | GRASP2 nonQTL |
| Waist circumfence | 4.5×10-4 | 25673412 (PMID) | Supplemental file |
Trans-association with RUXF supports role of splicing in Alzheimer's disease
This locus is discussed in the main paper. 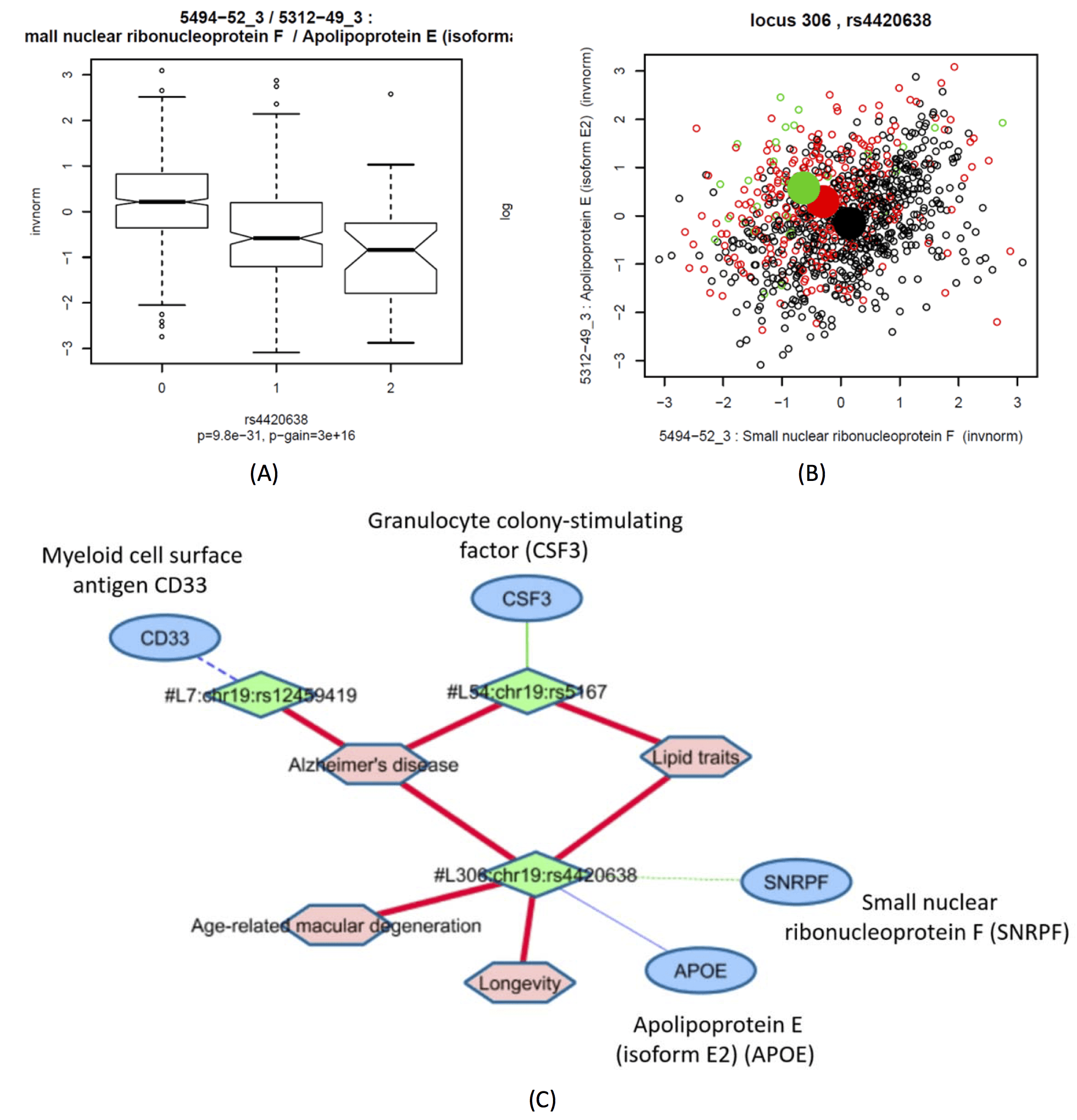
Boxplot of the ratio between Small Nuclear Ribonucleoprotein F (SNRPF aka RUXF) and Apolipoprotein E (isoform E2) with rs4420638 (Locus 306); Scatterplot of the ratio between SNRPF and APOE-2, coloured by genotype (black = major allele homozygotes, red = heterozygotes, green = minor allele homozygotes; means by genotype = large filled circles); subnetwork showing edges to three identified pQTLs at the APOE locus.
The information gathered here is a result of an attempt to keep track of all interesting information that we encountered while investigating these loci. Please bear in mind that the annotation given here is neither complete nor free of errors, and that all information provided here should be confirmed by additional literature research before being used as a basis for firm conclusions or further experiments.



























