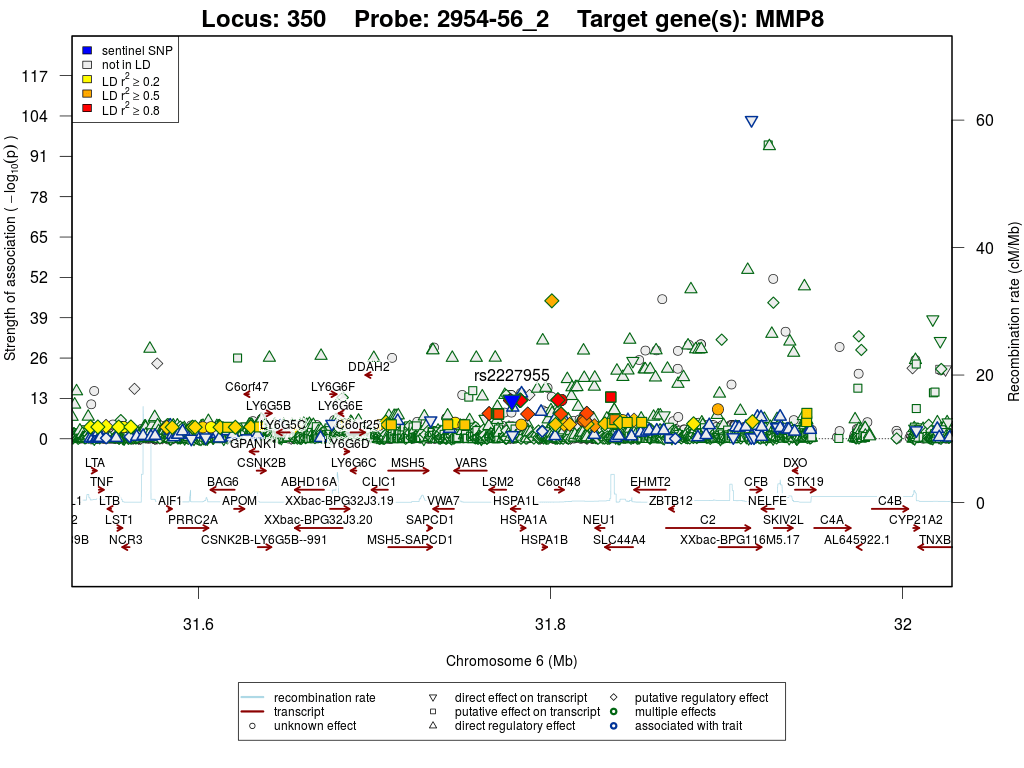
Locus annotations
Locus 350
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MMP-8 | trans | Discovery | rs2227955 | 6:31,778,077 | T/G | 0.02 | 997 | 1.024 | 0.139 | 4.2×10-13 | 1.390 | 1×10-13 | 4.5×10-11 |
| C34 gp41 HIV Fragment | trans | Discovery | rs2227955 | 6:31,778,077 | T/G | 0.02 | 997 | 0.848 | 0.143 | 3.8×10-9 | 1.230 | 2.2×10-8 | 3.5×10-5 |
Regional association plots
Neutrophil collagenase (MMP-8)
gp41 C34 peptide, HIV (C34 gp41 HIV Fragment)
Boxplots and histograms for top associations
Neutrophil collagenase (MMP-8)
| Target (abbrv.) | MMP-8 |
| Target (full name) | Neutrophil collagenase |
| Somalogic ID (Sequence ID) | SL000526 (2954-56_2) |
| Entrez Gene Symbol | MMP8 |
| UniProt ID | P22894 |
| UniProt Comment |
|
| Targeted by drugs (based on IPA annotation) |
|
| Biomarker applications (based on IPA annotation) |
|
gp41 C34 peptide, HIV (C34 gp41 HIV Fragment)
| Target (abbrv.) | C34 gp41 HIV Fragment |
| Target (full name) | gp41 C34 peptide, HIV |
| Somalogic ID (Sequence ID) | SL016148 (4792-51_2) |
| Entrez Gene Symbol | Human-virus |
| UniProt ID | Q70626 |
| UniProt Comment |
|
All locus annotations are based on the sentinel SNP (rs2227955) and 7 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by |
|
| Potentially regulated genes |
Results from other genome-wide association studies
Warning: count(): Parameter must be an array or an object that implements Countable in /var/www/html/pGWAS/locuscard.php on line 365
No associations available.