Locus annotations
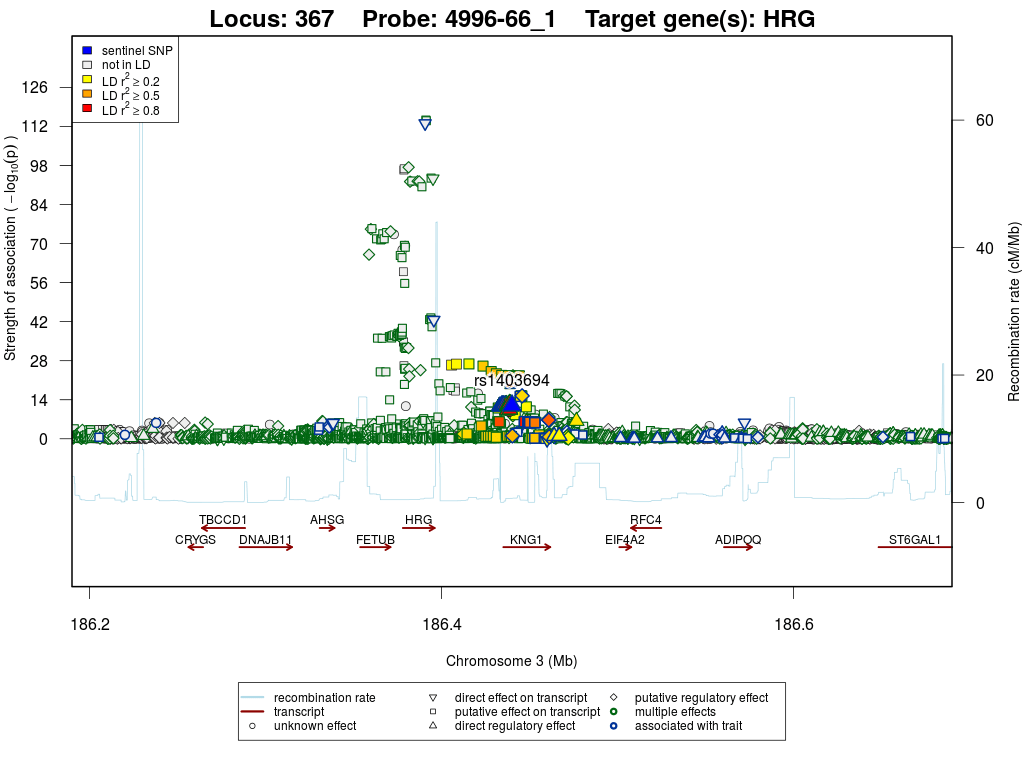
Locus 367
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HRG | cis | Discovery | rs1403694 | 3:186,439,998 | T/C | 0.39 | 994 | 0.320 | 0.044 | 1.2×10-12 | 1.070 | 3.8×10-12 | 8.6×10-12 |
| HRG | cis | Replication | rs1050274 | 3:186,435,370 | G/A | 0.41 | 337 | 0.245 | 0.072 | 0.001 | 1.060 | 0.026 | 0.001 |
Regional association plots
Histidine-rich glycoprotein (HRG)
Boxplots and histograms for top associations
Histidine-rich glycoprotein (HRG)
| Target (abbrv.) | HRG |
| Target (full name) | Histidine-rich glycoprotein |
| Somalogic ID (Sequence ID) | SL006448 (4996-66_1) |
| Entrez Gene Symbol | HRG |
| UniProt ID | P04196 |
| UniProt Comment |
|
| Reactome |
|
All locus annotations are based on the sentinel SNP (rs1403694) and 20 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by |
|
| Potentially regulated genes | |
| eQTL genes |
|
Results from other genome-wide association studies
| Trait | P | Study | Source |
|---|---|---|---|
| Methylation levels at chr3:187917754-187917804 [hg18 coord, probe cg12454167] in Temporal cortex | 8.4×10-21 | 20485568 (PMID) | GRASP2 meQTL |
| Methylation levels at chr3:187917754-187917804 [hg18 coord, probe cg12454167] in Caudal pons | 2.4×10-18 | 20485568 (PMID) | GRASP2 meQTL |
| Methylation levels at chr3:187917754-187917804 [hg18 coord, probe cg12454167] in Frontal cortex | 3.1×10-16 | 20485568 (PMID) | GRASP2 meQTL |
| Activated partial thromboplastin time | 9.5×10-14 | 23188048 (PMID) | GRASP2 nonQTL |
| Adiponectin levels | 1.5×10-9 | 22479202 (PMID) | GRASP2 nonQTL |
| Methylation levels at chr3:187917754-187917804 [hg18 coord, probe cg12454167] in Cerebellum | 2.6×10-9 | 20485568 (PMID) | GRASP2 meQTL |
| bradykinin, des-arg(9) | 2.2×10-5 | 24816252 (PMID) | gwas.eu via SNiPA |
| Gene expression of KNG1 in normal prepouch ileum | 1.1×10-4 | 23474282 (PMID) | GRASP2 eQTL |
| Serum creatinine | 3.7×10-4 | 20383146 (PMID) | GRASP2 nonQTL |