Locus 49
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Siglec-9 | cis | Discovery | rs2691273 | 19:51,596,136 | A/G | 0.46 | 993 | 0.764 | 0.039 | 3.1×10-73 | 4.930 | 1.3×10-81 | 1.1×10-89 |
| Siglec-9 | cis | Replication | rs2569428 | 19:51,608,796 | G/A | 0.42 | 337 | 0.517 | 0.066 | 8.1×10-14 | 3.150 | 9×10-16 | 5.2×10-14 |
Regional association plots
Sialic acid-binding Ig-like lectin 9 (Siglec-9)
Boxplots and histograms for top associations
Sialic acid-binding Ig-like lectin 9 (Siglec-9)
| Target (abbrv.) | Siglec-9 |
| Target (full name) | Sialic acid-binding Ig-like lectin 9 |
| Somalogic ID (Sequence ID) | SL005219 (3007-7_2) |
| Entrez Gene Symbol | SIGLEC9 |
| UniProt ID | Q9Y336 |
| UniProt Comment |
|
| Pathway Studio |
|
All locus annotations are based on the sentinel SNP (rs2691273) and 1 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by |
|
| eQTL genes |
|
Results from other genome-wide association studies
Warning: count(): Parameter must be an array or an object that implements Countable in /var/www/html/pGWAS/locuscard.php on line 365
No associations available.
An example of a genetically determined epitope effect.
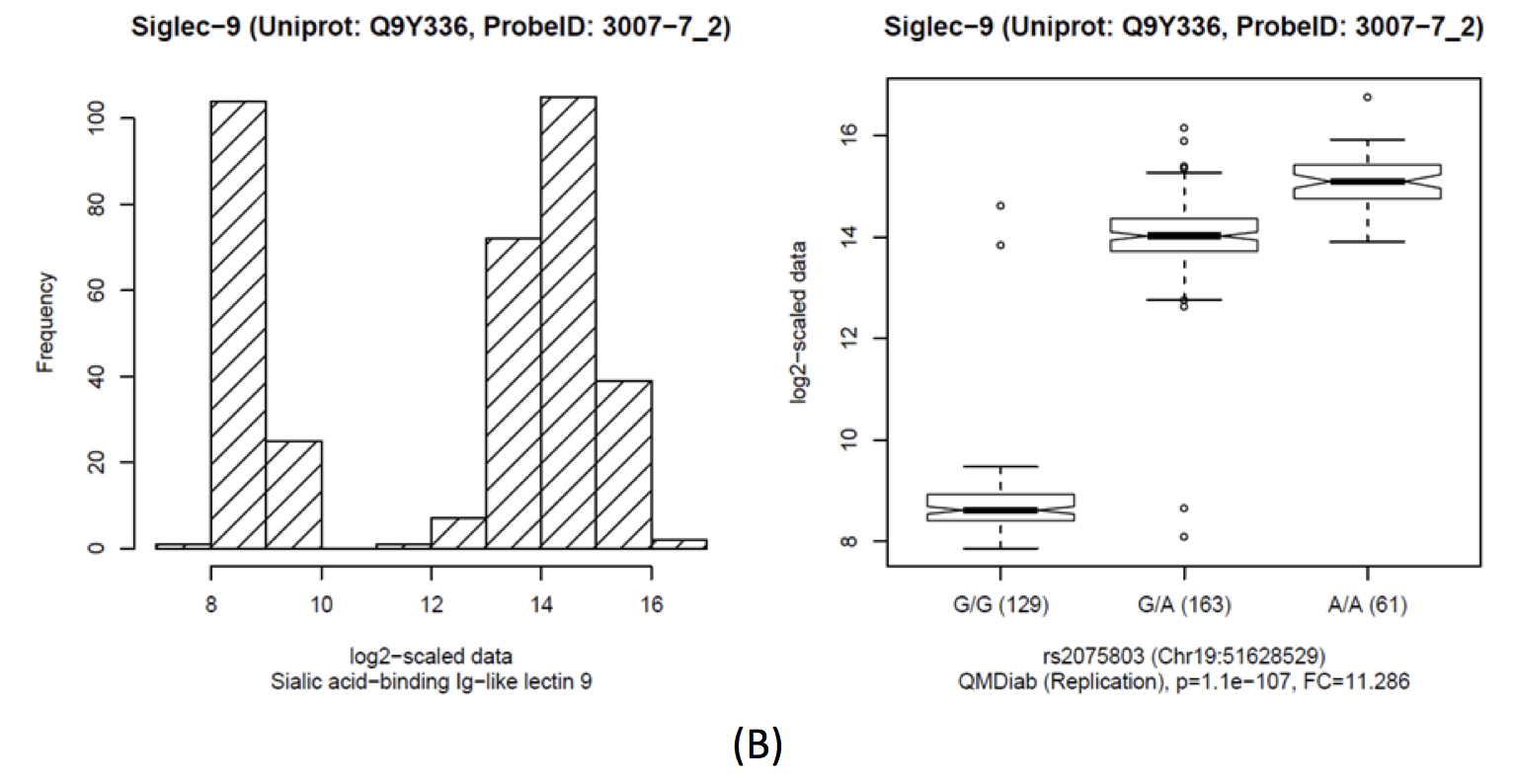
This locus harbours a replicated cis-pQTL with Sialic acid-binding Ig-like lectin 9 (SIGLEC9). SIGLEC9 levels display a strong bimodal distribution that replicates in QMDiab. Imputed data suggests two linked causative SNPs: rs2075803 (K100E) and rs2258983 (A315E) (r2=0.39 with rs2691273). These two variants were experimentally shown not to impact the gene function [PubMed]. rs2075803 (K100E) is located in the carbohydrate recognition domain of SIGLEC9. rs2075803 (K100E) is located in the carbohydrate recognition domain of SIGLEC9 and hence accessible to aptamer binding. Interesting enough, when excluding samples with very low SIGLEC9 levels, a functionally very convincing trans-association with rs10935480 (Locus 52) can be uncovered (p=7.5×10-6). The SIGLEC9 distribution is highly bi-modal, with SIGLEC9 levels of rs2075803-G homozygotes three orders of magnitude lower than those of carriers of the rs2075803-A allele. Interestingly, when excluding samples with low SIGLEC9 levels, a functionally relevant trans-association with rs10935480 (Locus 52) is found (p=7.5×10-6). rs10935480 is a replicated trans-association with Vascular endothelial growth factor receptor 3 (FLT4, aka VEGF sR3). The SNP is located near the ST3 Beta-Galactoside Alpha-2,3-Sialyltransferase 6 (ST3GAL6) gene, linking genetic variance at ST3GAL6 with FLT4 and SIGLEC9. 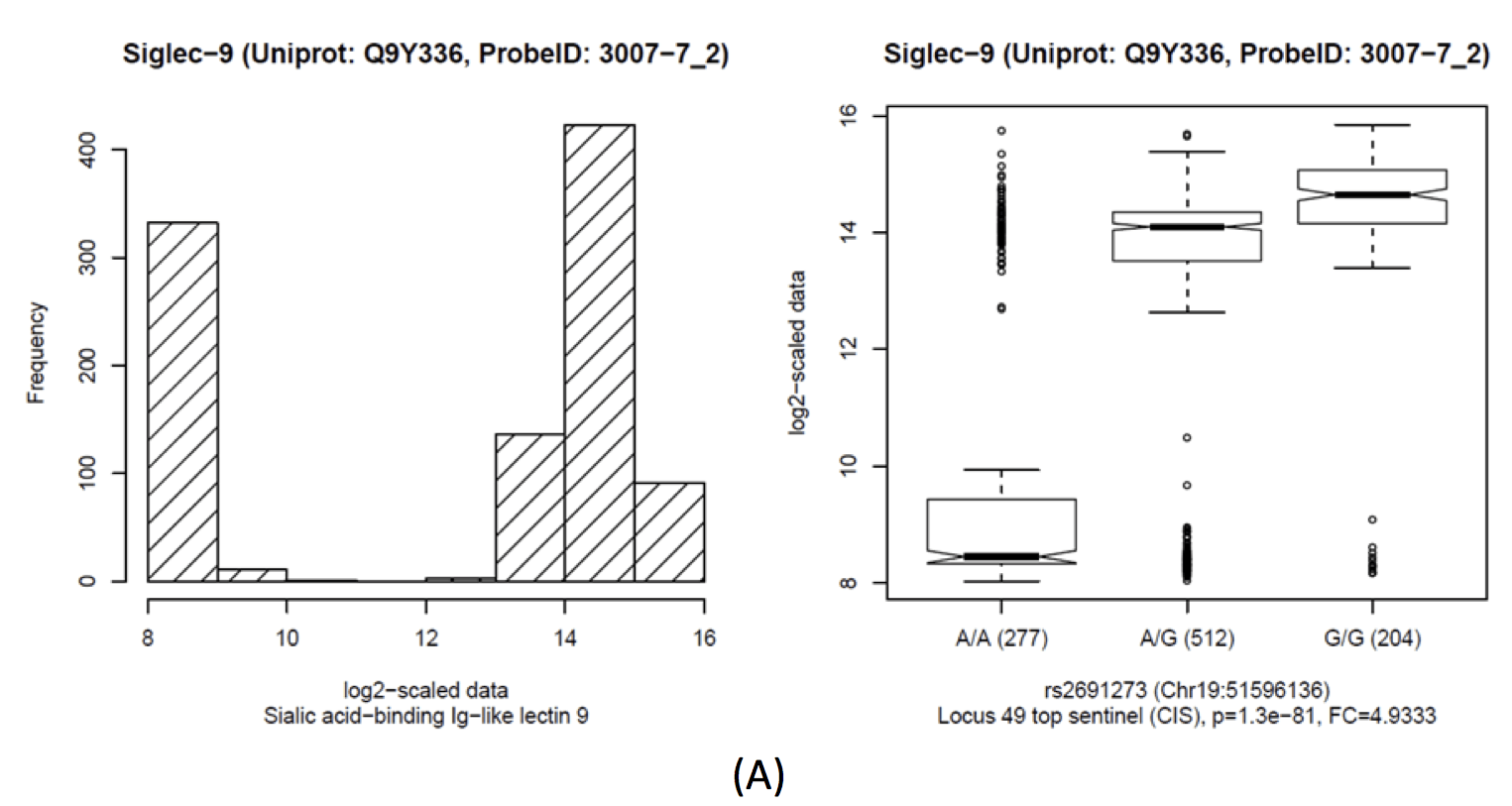
The information gathered here is a result of an attempt to keep track of all interesting information that we encountered while investigating these loci. Please bear in mind that the annotation given here is neither complete nor free of errors, and that all information provided here should be confirmed by additional literature research before being used as a basis for firm conclusions or further experiments.














