Locus 50
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CPNE1 | cis | Discovery | rs2425143 | 20:34,350,666 | C/T | 0.10 | 997 | -1.206 | 0.061 | 3.6×10-73 | -2.140 | 1.6×10-95 | 2×10-57 |
| CPNE1 | cis | Replication | rs12481228 | 20:34,218,673 | G/C | 0.15 | 337 | -0.877 | 0.045 | 2.2×10-56 | -2.510 | 6.7×10-58 | 2×10-32 |
Regional association plots
Copine-1 (CPNE1)
Boxplots and histograms for top associations
Copine-1 (CPNE1)
| Target (abbrv.) | CPNE1 |
| Target (full name) | Copine-1 |
| Somalogic ID (Sequence ID) | SL011808 (5346-24_3) |
| Entrez Gene Symbol | CPNE1 |
| UniProt ID | Q99829 |
| UniProt Comment |
|
| Reactome |
|
All locus annotations are based on the sentinel SNP (rs2425143) and 167 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by |
|
| Potentially regulated genes |
|
| eQTL genes |
|
Results from other genome-wide association studies
| Trait | P | Study | Source |
|---|---|---|---|
| Gene expression of CPNE1 in peripheral blood monocytes | 1.4×10-157 | 20502693 (PMID) | GRASP2 eQTL |
| Gene expression of RBM12 in blood | 3.9×10-115 | 21829388 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (probeID ILMN_1670841) in whole blood | 8×10-68 | 22692066 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (probeID ILMN_1670841) in breast tumors | 9.1×10-54 | 22522925 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (probeID ILMN_2307025) in breast tumors | 3.1×10-50 | 22522925 (PMID) | GRASP2 eQTL |
| CPNE1 gene expression | 2.3×10-46 | 21798870 (PMID) | GRASP2 eQTL |
| Gene expression of SCAND1 in blood | 5.6×10-46 | 21829388 (PMID) | GRASP2 eQTL |
| Gene expression | 5.6×10-38 | 20881960 (PMID) | GRASP2 nonQTL |
| cg23713742 (chr20:34202745) | 8.5×10-35 | 10.1101/033084 (DOI) | BIOS QTL cis-meQTLs |
| Height | 2.7×10-34 | 25282103 (PMID) | Supplemental file |
| Gene expression of CPNE1 [probe 206918_s_at] in lymphoblastoid cell lines | 1.3×10-33 | 17873877 (PMID) | GRASP2 eQTL |
| cg00719668 (chr20:34541037) | 3.8×10-31 | 10.1101/033084 (DOI) | BIOS QTL cis-meQTLs |
| Gene expression of CPNE1 in CEU-CHB-JPT-YRI lymphoblastoid cell lines | 2.3×10-28 | 17873874 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in CEU-CHB-JPT lymphoblastoid cell lines | 3.6×10-23 | 17873874 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 [probe ILMN_6266] in osteoblasts treated with PGE2 | 2.6×10-20 | 21283786 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 [probe ILMN_6266] in osteoblasts treated with BMP2 | 7.9×10-17 | 21283786 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in lymphoblastoid cell lines | 1.4×10-16 | 18193047 (PMID) | GRASP2 eQTL |
| "Gene expression of CPNE1 [transcript NM_152928, probe A_23_P218726] in liver" | 2×10-16 | 21637794 (PMID) | GRASP2 eQTL |
| cg07042104 (chr20:34206975) | 2.6×10-16 | 10.1101/033084 (DOI) | BIOS QTL cis-meQTLs |
| Gene expression of CPNE1 in liver | 7.1×10-16 | 18462017 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 [probe ILMN_6266] in osteoblasts treated with dexamethasone | 1.5×10-15 | 21283786 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (ENSG00000214078) in dendritic cells treated with Mycobacterium tuberculosis | 1.5×10-15 | 22233810 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in normal prepouch ileum | 4.7×10-13 | 23474282 (PMID) | GRASP2 eQTL |
| Gene expression of RBM12 in Fibroblasts | 8×10-13 | 19644074 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in CEU lymphoblastoid cell lines | 1.7×10-12 | 17873874 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (ENSG00000214078) in dendritic cells | 2.7×10-12 | 22233810 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 [probe ILMN_6266] in osteoblasts | 5.6×10-12 | 19654370 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 [probe ILMN_6266] in untreated osteoblasts | 5.6×10-12 | 21283786 (PMID) | GRASP2 eQTL |
| Gene expression of RBM12 in Lymphoblastoid cell lines | 6.6×10-12 | 19644074 (PMID) | GRASP2 eQTL |
| Hip circumfence, adjusted for BMI | 3×10-11 | 25673412 (PMID) | Supplemental file |
| Differential exon level expression of CPNE1 [probe 3904125] in peripheral blood mononuclear cells | 2.4×10-10 | 19222302 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904139] in peripheral blood mononuclear cells | 1.4×10-9 | 19222302 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904130] in peripheral blood mononuclear cells | 3.2×10-9 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (probeID ILMN_2307025) in temporal cortex in Alzheimer's disease cases and controls | 1×10-8 | 22685416 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in JPT lymphoblastoid cell lines | 2×10-8 | 17873874 (PMID) | GRASP2 eQTL |
| Triglycerides | 7.6×10-8 | 24097068 (PMID) | Supplemental file |
| Differential exon level expression of CPNE1 [probe 3904131] in peripheral blood mononuclear cells | 8.8×10-8 | 19222302 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904143] in peripheral blood mononuclear cells | 1.6×10-7 | 19222302 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904126] in peripheral blood mononuclear cells | 2.5×10-7 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in CHB lymphoblastoid cell lines | 9×10-7 | 17873874 (PMID) | GRASP2 eQTL |
| Total cholesterol | 1×10-6 | 20686565 (PMID) | GRASP2 nonQTL |
| cg08161081 (chr20:34327442) | 1.3×10-6 | 10.1101/033084 (DOI) | BIOS QTL cis-meQTLs |
| Differential exon level expression of CPNE1 [probe 3904136] in peripheral blood mononuclear cells | 2.2×10-6 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 (probeID ILMN_2307025) in cerebellum in Alzheimer's disease cases and controls | 2.3×10-6 | 22685416 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904122] in peripheral blood mononuclear cells | 2.5×10-6 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of TMEM48 in peripheral blood monocytes | 2.5×10-6 | 20502693 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904123] in peripheral blood mononuclear cells | 2.9×10-6 | 19222302 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904138] in peripheral blood mononuclear cells | 4.1×10-6 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of FXYD1 in peripheral blood monocytes | 4.8×10-6 | 20502693 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904121] in peripheral blood mononuclear cells | 6.3×10-6 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of RBM12 in T cells | 6.5×10-6 | 19644074 (PMID) | GRASP2 eQTL |
| Hip circumfence | 9.1×10-6 | 25673412 (PMID) | Supplemental file |
| Gene expression of CPNE1 [probe 3904119] in brain cortex | 1.9×10-5 | 19222302 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904131] in brain cortex | 2.4×10-5 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 in CD4+ lymphocytes | 3.1×10-5 | 20833654 (PMID) | GRASP2 eQTL |
| Differential exon level expression of CPNE1 [probe 3904139] in brain cortex | 5.1×10-5 | 19222302 (PMID) | GRASP2 eQTL |
| LDL cholesterol | 5.9×10-5 | 24097068 (PMID) | Supplemental file |
| Differential exon level expression of CPNE1 [probe 3904136] in brain cortex | 6.4×10-5 | 19222302 (PMID) | GRASP2 eQTL |
| Gene expression of CPNE1 [probe 3904119] in peripheral blood mononuclear cells | 2.5×10-4 | 19222302 (PMID) | GRASP2 eQTL |
Near total protein ablation in minor allele homozygotes, confirmed by allele specific RNA expression analysis.
Cis-association with Copine-1 (CPNE1) – see Supplemental Figure 5 below. 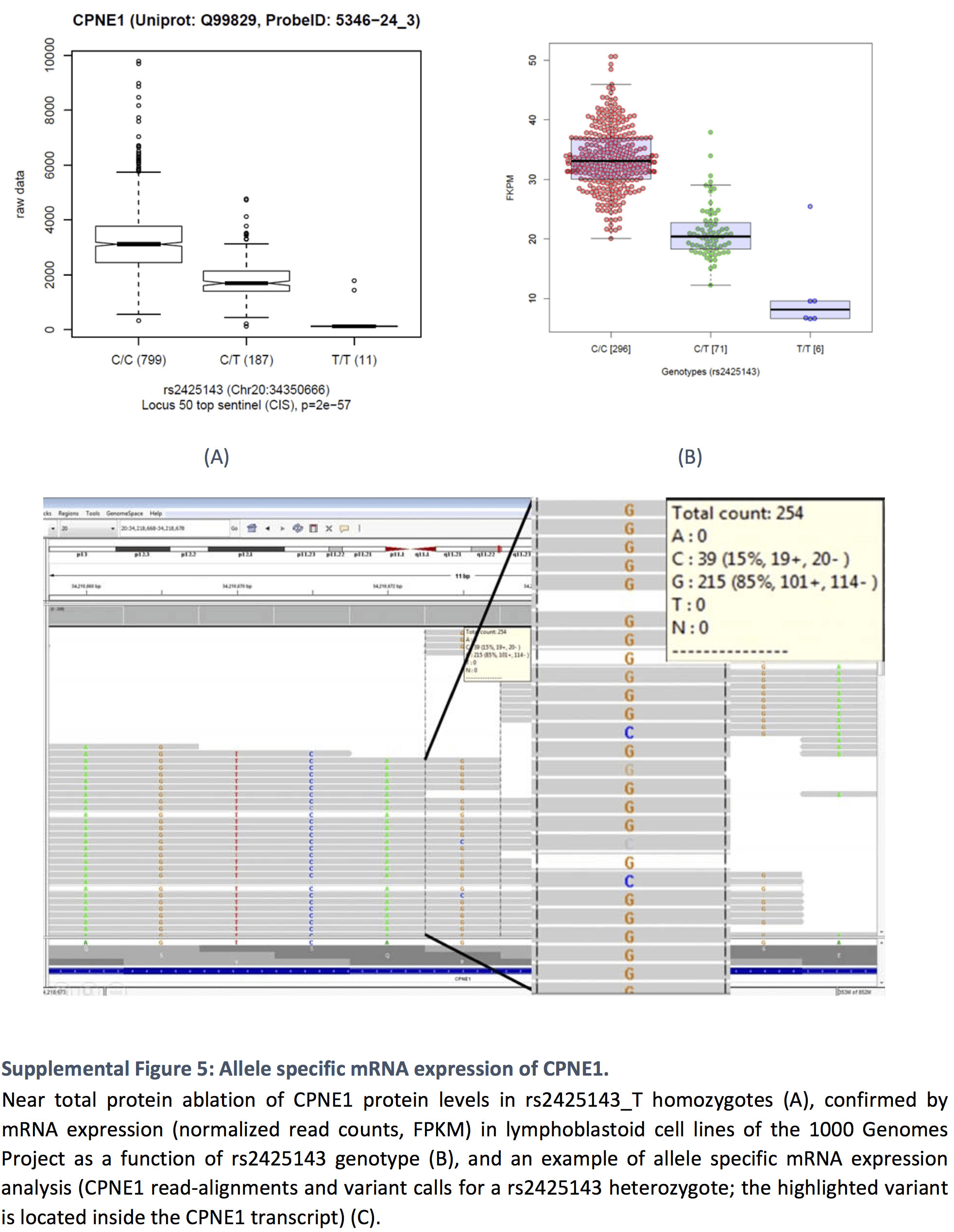
The information gathered here is a result of an attempt to keep track of all interesting information that we encountered while investigating these loci. Please bear in mind that the annotation given here is neither complete nor free of errors, and that all information provided here should be confirmed by additional literature research before being used as a basis for firm conclusions or further experiments.














