Locus 67
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Haptoglobin, Mixed Type | cis | Discovery | rs9302635 | 16:72,144,174 | T/C | 0.18 | 997 | 0.876 | 0.053 | 6.3×10-54 | 1.530 | 1.7×10-43 | 5.3×10-56 |
| Haptoglobin, Mixed Type | cis | Replication | rs9302635 | 16:72,144,174 | A/G | 0.17 | 337 | 0.859 | 0.091 | 6.2×10-19 | 1.900 | 6.5×10-12 | 7.7×10-22 |
| Ferritin | trans | Discovery | rs9302635 | 16:72,144,174 | T/C | 0.18 | 997 | 0.372 | 0.058 | 2.8×10-10 | 1.150 | 2.4×10-6 | 0.043 |
| Ferritin | trans | Replication | rs9302635 | 16:72,144,174 | A/G | 0.17 | 337 | 0.571 | 0.098 | 1.4×10-8 | 1.240 | 2.5×10-5 | 0.873 |
Regional association plots
Haptoglobin (Haptoglobin, Mixed Type)
Ferritin
Boxplots and histograms for top associations
Haptoglobin (Haptoglobin, Mixed Type)
| Target (abbrv.) | Haptoglobin, Mixed Type |
| Target (full name) | Haptoglobin |
| Somalogic ID (Sequence ID) | SL000437 (3054-3_2) |
| Entrez Gene Symbol | HP |
| UniProt ID | P00738 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
| Reactome |
|
Ferritin
| Target (abbrv.) | Ferritin |
| Target (full name) | Ferritin |
| Somalogic ID (Sequence ID) | SL000420 (3031-66_1) |
| Entrez Gene Symbol | FTH1, FTL |
| UniProt ID | P02794, P02792 |
| UniProt Comment |
|
All locus annotations are based on the sentinel SNP (rs9302635) and 6 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by | |
| eQTL genes |
Results from other genome-wide association studies
| Trait | P | Study | Source |
|---|---|---|---|
| Gene expression of HP in peripheral blood monocytes | 2.2×10-41 | 20502693 (PMID) | GRASP2 eQTL |
| LDL cholesterol | 2×10-15 | 24097068 (PMID) | Supplemental file |
| Triglycerides | 1.4×10-14 | 24097068 (PMID) | Supplemental file |
| Total cholesterol | 5.1×10-9 | 20686565 (PMID) | GRASP2 nonQTL |
| gamma-glutamyltyrosine | 9.2×10-5 | 24816252 (PMID) | gwas.eu via SNiPA |
New cholesterol association.
Using http://genenetwork.nl/biosqtlbrowser/ we identified different effects on transcripts and methylome:
Locus 46: rs217181 => cis-eQTL Exon-Level 2.3×10-40 chr_16_72108183_72108650 with HPR (effect on the HP related gene)
Locus 67: rs9302635 => cis-eQTL Exon-ratio 1.1×10-32 with ENSG00000257017_16_72094011_72094954 positive score with HP, 8.9×10-41 with ENSG00000257017_16_72092153_72092408 negative score with HP (effect on splice variants)
Locus 86: rs2000999 => trans-meQTL 2.8×10-7 with cg23809679 on chr4:71,554,149 (putative effect on distant gene, possibly UTP3)
The following associations are with HPR! 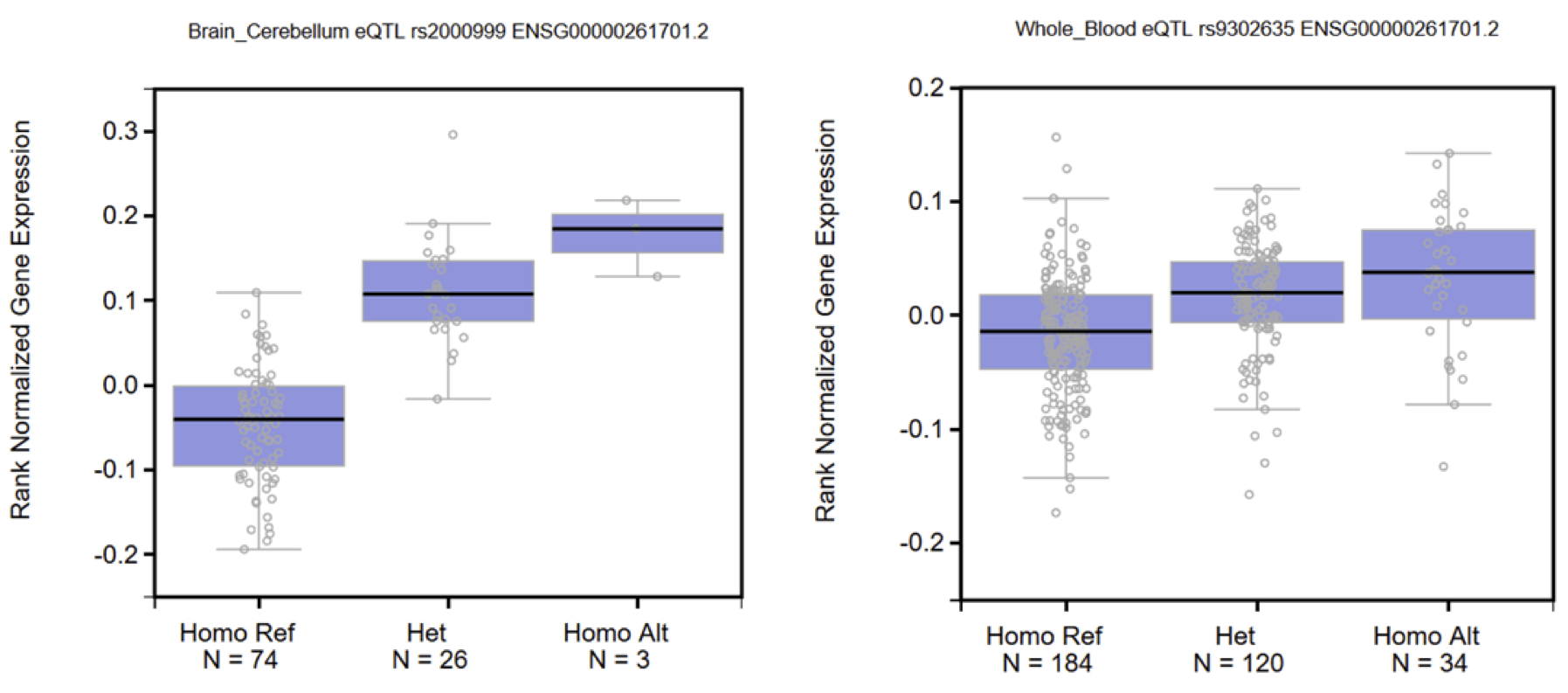
SNPs rs217181 and rs2000999 were associated in cis with haptoglobin (HP) and in trans with hemoglobin (HBA1/HBB) and heparin cofactor 2 (SERPIND1, also HC II). The effect sizes of both variants were comparable (FC=1.58 per copy of the minor allele for rs217181 and FC=-1.48 for rs2000999 on haptoglobin levels). The effect sizes of both "risk" alleles are additive and lead to a similar effect size of the association with the minor allele-copy difference between both SNPs (FC=1.41). The association of this difference strengthens the association at this locus with haptoglobin by 27 orders of magnitude.
Given that the effects of both variants are of similar strength and opposite direction, and that the minor allele frequencies appear to be similar (36 and 38 minor allele homozygotes in this study, resp.), one can expect the association signal for rs217181 with LDL and total cholesterol to be similar as for rs2000999, but with opposite direction. This was confirmed by a lookup in the association data from the Global Lipids Genetics Consortium Results [PubMed] : Both SNPs associate with a reverse sign with LDL (rs217181-T decreases LDL with beta=-0.046 and p=1.1×10-25; rs2000999-A increases LDL with beta=0.065 and p=4.2×10-41), total cholesterol (similar association strength), and triglycerides (weak association signal). There is thus one low frequency variant that increases LDL and one low frequency variant that decreases LDL. This is an example for a drug response case (see Plenge et al. [PubMed]). The rs217181-LDL association was never reported.
Froguel et al. [PubMed] identified rs2000999 as a strong genetic determinant of circulating haptoglobin levels.
This SNP was also reported as a lipid risk variant, associated with LDL cholesterol and with total cholesterol [PubMed]. rs217181 was reported in association with glycoprotein acetyls (GlycA, mainly a1-acid glycoprotein, p=1.46×10-36) [PubMed], and replicated by Frazier-Wood et al. (Circulation. 2015;131:AP361). GlycA is a new NMR-derived plasma marker of inflammation.
Also encoded at the locus is haptoglobin-related protein (HPR). HPR derived from HP by gene duplication. SNP rs2000999 is an intronic SNP in HPR and rs217181 is located 2,857 bases downstream of HPR. RefSeq states: "This gene encodes a haptoglobin-related protein that binds hemoglobin as efficiently as haptoglobin. Unlike haptoglobin, plasma concentration of this protein is unaffected in patients with sickle cell anemia and extensive intravascular hemolysis, suggesting a difference in binding between haptoglobin-hemoglobin and haptoglobin-related protein-hemoglobin complexes to CD163, the hemoglobin scavenger receptor. This protein may also be a clinically important predictor of recurrence of breast cancer."
Uniprot states "Primate-specific plasma protein associated with apolipoprotein L-I (apoL-I)-containing high-density lipoprotein (HDL). This HDL particle, termed trypanosome lytic factor-1 (TLF-1), mediates human innate immune protection against many species of African trypanosomes." This paper refers to this review [PubMed], which may also explain the association of the rs2000999 variant with LDL and total cholesterol: "TLF1 is the densest fraction of high density lipoprotein particles (known as fraction 3 or HDL3), which contain a lipid core with an outer hydrophilic layer of phospholipids, cholesterol and several apolipoproteins, including the major component apolipoprotein A1 (APOA1)" [PubMed].
Heparin cofactor 2 is a plasma serine proteinase inhibitor (serpin) that inhibits the coagulant proteinase alpha-thrombin. Peptides at the N-terminal of SERPIND1 have chemotactic activity for both monocytes and neutrophils. 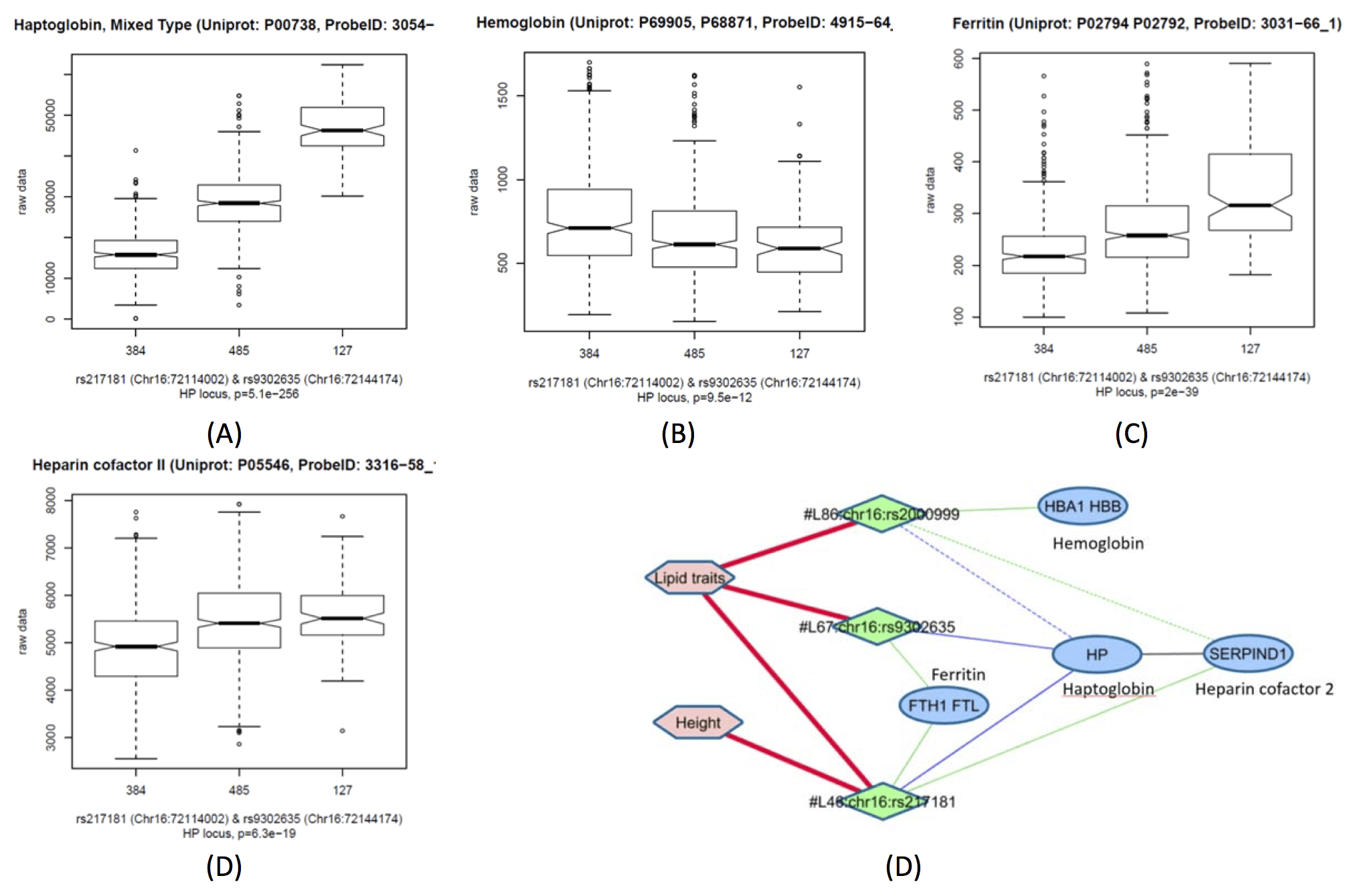
Association network at the haptoglobin locus. Boxplots of Haptoglobin (HP), Hemoglobin (HBA1/HBB), Ferritin (FTH1/FTL), and Heparin cofactor 2 (SERPIND1) protein levels (outliers >4s.d. were removed) as a function of the minor allele copy number of rs217181 (A-D). Sub-network depeicting the haptoglobin (HP) locus, with three independent genetic associations that link four proteins and GWAS associations with lipid traits. The association with HP is a cis-pQTL, all others are trans-pQTLs.
The information gathered here is a result of an attempt to keep track of all interesting information that we encountered while investigating these loci. Please bear in mind that the annotation given here is neither complete nor free of errors, and that all information provided here should be confirmed by additional literature research before being used as a basis for firm conclusions or further experiments.



























