Locus 69
Top associations per target
| Target | cis/trans | Study | SNP | SNP location | Maj/min allele | MAF | N | βinv | seinv | Pinv | fclog | Plog | Praw |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cadherin-5 | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 1.108 | 0.068 | 6.1×10-53 | 1.240 | 1.2×10-55 | 1.3×10-52 |
| Cadherin-5 | trans | Replication | rs8176693 | 9:136,137,657 | G/A | 0.16 | 337 | 0.933 | 0.059 | 2.7×10-42 | 1.240 | 2.9×10-42 | 3.5×10-40 |
| sTie-1 | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 1.099 | 0.069 | 1.7×10-51 | 1.210 | 5.8×10-51 | 1×10-57 |
| sTie-1 | trans | Replication | rs8176743 | 9:136,131,415 | G/A | 0.17 | 337 | 0.804 | 0.068 | 3.5×10-27 | 1.220 | 10×10-26 | 1.2×10-25 |
| sTie-2 | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.906 | 0.070 | 1.1×10-35 | 1.130 | 2.2×10-36 | 8.7×10-38 |
| sTie-2 | trans | Replication | rs8176693 | 9:136,137,657 | G/A | 0.16 | 337 | 0.496 | 0.075 | 2×10-10 | 1.090 | 5.6×10-10 | 3.4×10-9 |
| Endoglin | cis | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.629 | 0.074 | 5.6×10-17 | 1.130 | 1.4×10-16 | 3.9×10-18 |
| Endoglin | cis | Replication | rs8176743 | 9:136,131,415 | G/A | 0.17 | 337 | 0.260 | 0.072 | 3.4×10-4 | 1.100 | 0.002 | 0.092 |
| BCAM | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.506 | 0.074 | 1.5×10-11 | 1.090 | 7.7×10-8 | 0.08 |
| BCAM | trans | Replication | rs8176693 | 9:136,137,657 | G/A | 0.16 | 337 | 0.282 | 0.069 | 5.8×10-5 | 1.070 | 2.7×10-4 | 0.001 |
| CD109 | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.509 | 0.075 | 1.7×10-11 | 1.120 | 1.8×10-10 | 9.2×10-9 |
| CD109 | trans | Replication | rs8176693 | 9:136,137,657 | G/A | 0.16 | 337 | 0.260 | 0.074 | 4.8×10-4 | 1.110 | 3.1×10-4 | 0.001 |
| VEGF sR3 | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.492 | 0.075 | 7.9×10-11 | 1.110 | 1.2×10-9 | 9.9×10-11 |
| VEGF sR3 | trans | Replication | rs77693339 | 9:136,128,558 | A/G | 0.16 | 337 | 0.237 | 0.087 | 0.007 | 1.060 | 0.009 | 0.01 |
| TCCR | trans | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.492 | 0.075 | 8.3×10-11 | 1.130 | 1.9×10-9 | 0.004 |
| TCCR | trans | Replication | rs8176743 | 9:136,131,415 | G/A | 0.17 | 337 | 0.221 | 0.093 | 0.017 | 1.030 | 0.278 | 0.799 |
| Notch 1 | cis | Discovery | rs8176749 | 9:136,131,188 | C/T | 0.09 | 995 | 0.448 | 0.072 | 6×10-10 | 1.050 | 2.3×10-9 | 6.4×10-10 |
| Notch 1 | cis | Replication | rs8176743 | 9:136,131,415 | G/A | 0.17 | 337 | 0.235 | 0.051 | 5.3×10-6 | 1.040 | 3.1×10-6 | 8.9×10-6 |
Regional association plots
Cadherin-5
Tyrosine-protein kinase receptor Tie-1, soluble (sTie-1)
Angiopoietin-1 receptor, soluble (sTie-2)
Endoglin
Basal Cell Adhesion Molecule (BCAM)
CD109 antigen (CD109)
Vascular endothelial growth factor receptor 3 (VEGF sR3)
Interleukin-27 receptor subunit alpha (TCCR)
Neurogenic locus notch homolog protein 1 (Notch 1)
Boxplots and histograms for top associations
Cadherin-5
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Tyrosine-protein kinase receptor Tie-1, soluble (sTie-1)
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Angiopoietin-1 receptor, soluble (sTie-2)
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Endoglin
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Basal Cell Adhesion Molecule (BCAM)
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
CD109 antigen (CD109)
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Vascular endothelial growth factor receptor 3 (VEGF sR3)
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Interleukin-27 receptor subunit alpha (TCCR)
| inverse-normalized probe levels | log2 transformed probe levels | raw probe levels | |
|---|---|---|---|
| Discovery study |


|


|


|
| Replication study |


|


|


|
Neurogenic locus notch homolog protein 1 (Notch 1)
Cadherin-5
| Target (abbrv.) | Cadherin-5 |
| Target (full name) | Cadherin-5 |
| Somalogic ID (Sequence ID) | SL002081 (2819-23_2) |
| Entrez Gene Symbol | CDH5 |
| UniProt ID | P33151 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
| Pathway Interaction Database |
|
| Reactome |
|
| Pathway Studio |
|
Tyrosine-protein kinase receptor Tie-1, soluble (sTie-1)
| Target (abbrv.) | sTie-1 |
| Target (full name) | Tyrosine-protein kinase receptor Tie-1, soluble |
| Somalogic ID (Sequence ID) | SL003199 (2844-53_2) |
| Entrez Gene Symbol | TIE1 |
| UniProt ID | P35590 |
| UniProt Comment |
|
Angiopoietin-1 receptor, soluble (sTie-2)
| Target (abbrv.) | sTie-2 |
| Target (full name) | Angiopoietin-1 receptor, soluble |
| Somalogic ID (Sequence ID) | SL003200 (3773-15_4) |
| Entrez Gene Symbol | TEK |
| UniProt ID | Q02763 |
| UniProt Comment |
|
| Targeted by drugs (based on IPA annotation) |
|
| Biomarker applications (based on IPA annotation) |
|
| Wiki Pathways |
|
| Pathway Interaction Database |
|
| Reactome |
|
| Pathway Studio |
|
Endoglin
| Target (abbrv.) | Endoglin |
| Target (full name) | Endoglin |
| Somalogic ID (Sequence ID) | SL004482 (4908-6_1) |
| Entrez Gene Symbol | ENG |
| UniProt ID | P17813 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
| Wiki Pathways |
|
| Pathway Interaction Database |
|
Basal Cell Adhesion Molecule (BCAM)
| Target (abbrv.) | BCAM |
| Target (full name) | Basal Cell Adhesion Molecule |
| Somalogic ID (Sequence ID) | SL001902 (2816-50_2) |
| Entrez Gene Symbol | BCAM |
| UniProt ID | P50895 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
CD109 antigen (CD109)
| Target (abbrv.) | CD109 |
| Target (full name) | CD109 antigen |
| Somalogic ID (Sequence ID) | SL008773 (3290-50_2) |
| Entrez Gene Symbol | CD109 |
| UniProt ID | Q6YHK3 |
| UniProt Comment |
|
Vascular endothelial growth factor receptor 3 (VEGF sR3)
| Target (abbrv.) | VEGF sR3 |
| Target (full name) | Vascular endothelial growth factor receptor 3 |
| Somalogic ID (Sequence ID) | SL003322 (2358-19_2) |
| Entrez Gene Symbol | FLT4 |
| UniProt ID | P35916 |
| UniProt Comment |
|
| Targeted by drugs (based on IPA annotation) |
|
| Biomarker applications (based on IPA annotation) |
|
| Pathway Interaction Database |
|
| Reactome |
|
| Pathway Studio |
|
Interleukin-27 receptor subunit alpha (TCCR)
| Target (abbrv.) | TCCR |
| Target (full name) | Interleukin-27 receptor subunit alpha |
| Somalogic ID (Sequence ID) | SL005223 (5132-71_3) |
| Entrez Gene Symbol | IL27RA |
| UniProt ID | Q6UWB1 |
| UniProt Comment |
|
| Pathway Interaction Database |
|
| Pathway Studio |
|
Neurogenic locus notch homolog protein 1 (Notch 1)
| Target (abbrv.) | Notch 1 |
| Target (full name) | Neurogenic locus notch homolog protein 1 |
| Somalogic ID (Sequence ID) | SL005703 (5107-7_2) |
| Entrez Gene Symbol | NOTCH1 |
| UniProt ID | P46531 |
| UniProt Comment |
|
| Biomarker applications (based on IPA annotation) |
|
| Wiki Pathways |
|
| Pathway Interaction Database |
|
| Reactome |
|
| Pathway Studio |
|
All locus annotations are based on the sentinel SNP (rs8176749) and 24 proxy variant(s) that is/are in linkage disequilibrium r2 ≥ 0.8. Linkage disequilibrium is based on data from the 1000 Genomes Project, phase 3 version 5, European population and was retrieved using SNiPA's Block Annotation feature.
Download the detailed results of SNiPA's block annotation (PDF)
Linked genes
| Genes hit or close-by | |
| Potentially regulated genes |
|
| eQTL genes |
Results from other genome-wide association studies
| Trait | P | Study | Source |
|---|---|---|---|
| Coagulation factor levels | 2×10-138 | 23267103 (PMID) | GWAS Catalog via SNiPA |
| Tumor biomarkers | 7×10-105 | 23300138 (PMID) | GWAS Catalog via SNiPA |
| Plasma carcinoembryonic antigen (CEA) levels | 7.2×10-105 | 23300138 (PMID) | GRASP2 nonQTL |
| Metabolic syndrome domains (Multivariate analysis) | 6.1×10-75 | 22022282 (PMID) | GRASP2 nonQTL |
| von Willebrand factor (vWF) | 4.4×10-36 | 23267103 (PMID) | GRASP2 nonQTL |
| cg07241568 (chr9:136150847) | 9.8×10-31 | 10.1101/033084 (DOI) | BIOS QTL cis-meQTLs |
| Elevated serum carcinoembryonic antigen levels | 2×10-24 | 24941225 (PMID) | GWAS Catalog via SNiPA |
| Activated partial thromboplastin time | 2.9×10-23 | 22703881 (PMID) | GRASP2 nonQTL |
| High serum lipase activity | 1×10-22 | 25028398 (PMID) | GWAS Catalog via SNiPA |
| Hemoglobin (Hb) | 1.1×10-17 | 23222517 (PMID) | GRASP2 nonQTL |
| End-stage coagulation | 2×10-17 | 23381943 (PMID) | GWAS Catalog via SNiPA |
| Factor XIII antigen | 1.2×10-14 | 23381943 (PMID) | GRASP2 nonQTL |
| TNF-alpha (TNF-a) | 2×10-14 | 18464913 (PMID) | GRASP2 nonQTL |
| Red blood cell count (RBC) | 1.6×10-13 | 23222517 (PMID) | GRASP2 nonQTL |
| Urinary metabolites (H-NMR features) | 4×10-12 | 24586186 (PMID) | GWAS Catalog via SNiPA |
| LDL cholesterol | 2.2×10-11 | 24097068 (PMID) | Supplemental file |
| Intraocular pressure | 3×10-11 | 25173106 (PMID) | GWAS Catalog via SNiPA |
| Gene expression of MED22 in peripheral blood monocytes | 1.8×10-10 | 20502693 (PMID) | GRASP2 eQTL |
| Angiotensin-converting enzyme (ACE) activity | 2.9×10-10 | 20066004 (PMID) | GRASP2 nonQTL |
| Malaria | 3.7×10-8 | 23717212 (PMID) | GRASP2 nonQTL |
| Mean corpuscular hemoglobin concentration | 4×10-8 | 20139978 (PMID) | GWAS Catalog via SNiPA |
| Mean corpuscular hemoglobin concentration (MCHC) | 4.3×10-8 | 20139978 (PMID) | GRASP2 nonQTL |
| Hematocrit (Hct) | 5.4×10-8 | 23222517 (PMID) | GRASP2 nonQTL |
| Gene expression of ABO (probeID ILMN_1656979) in temporal cortex in Alzheimer's disease cases and controls | 7.1×10-8 | 22685416 (PMID) | GRASP2 eQTL |
| Circulating galectin-3 levels | 7.6×10-8 | 23056639 (PMID) | GRASP2 nonQTL |
| cg23778596 (chr7:149411474) | 9.8×10-8 | 10.1101/033084 (DOI) | BIOS QTL trans-meQTLs |
| Mean corpuscular volume (MCV) | 1.5×10-7 | 23222517 (PMID) | GRASP2 nonQTL |
| Gene expression of SURF6 in peripheral blood monocytes | 2.8×10-7 | 20502693 (PMID) | GRASP2 eQTL |
| Gene expression of ABO (probeID ILMN_1656979) in temporal cortex in Alzheimer's disease cases | 2×10-6 | 22685416 (PMID) | GRASP2 eQTL |
| Gene expression of ABO in CD4+ lymphocytes | 2.8×10-6 | 20833654 (PMID) | GRASP2 eQTL |
| Graves' disease | 4×10-6 | 23612905 (PMID) | GRASP2 nonQTL |
| Parkinson disease | 8.7×10-6 | pha002868 (dbGaP) | dbGaP via SNiPA |
| Parkinson's disease (PD) | 8.7×10-6 | 19915575 (PMID) | GRASP2 nonQTL |
| Gene expression of ABO (probeID ILMN_1656979) in cerebellum in Alzheimer's disease cases and controls | 1.1×10-5 | 22685416 (PMID) | GRASP2 eQTL |
| Gene expression of ABO in normal prepouch ileum | 1.2×10-5 | 23474282 (PMID) | GRASP2 eQTL |
| Gene expression of ABO (probeID ILMN_1656979) in cerebellum in Alzheimer's disease cases | 1.2×10-5 | 22685416 (PMID) | GRASP2 eQTL |
| HDL cholesterol | 2.1×10-5 | 24097068 (PMID) | Supplemental file |
| Grave's disease | 2.6×10-5 | 21841780 (PMID) | GRASP2 nonQTL |
| Esophageal squamous cell carcinoma (Esophageal cancer) | 7.1×10-5 | 23300138 (PMID) | GRASP2 nonQTL |
| Autism | 1.5×10-4 | 20663923 (PMID) | GRASP2 nonQTL |
| Chronic kidney disease | 4.8×10-4 | 20383146 (PMID) | GRASP2 nonQTL |
The pleiotropic ABO locus is discussed in the main paper.
This locus is discussed in the main paper. Here we report observations not mentioned in the paper: In Shin et al. [PubMed] SNP rs651007 associates with dipeptide levels (aspartylphenylalanine, p=2×10-7; X-14189, annotated as potential Leu-Ala, but Ser-Pro is also possible by mass alone, p=9.6×10-10; X14304, I/L-Ala or Ala-I/L?, p=1.8×10-8; X-14208, could be Cys-Met or Tyr-Ala or His-Pro or Ser-Phe by mass alone, p=1.5×10-7; X-14205; X14086: annotated as Thr-Glu, but mass is 25 too high; Val-Arg possible by mass alone). This SNP also associates in Raffler et al. [PubMed] (Urine NMR) with a signal at 2.0308 ppm (PM20308, p=5.1×10-20). It is also a marginal hit in CardioGram (7.1×10-8). He et al. [PubMed] report a genome wide association study of genetic loci that influence tumour biomarkers cancer antigen 19-9, carcinoembryonic antigen and α fetoprotein and their associations with cancer risk. The authors find strong association of SNP rs8176749 with carcinoembryonic antigen (CEA) levels – we find association with Cadherin-5. CD209 and MBL2 are both lectins. All associated ABO-associated proteins are glycoproteins, part of glycoprotein complex (LY96), or glycoprotein-binding (SELE), except for IL27RA (http://jcggdb.jp/rcmg/gpdb/index). According to Uniprot all proteins are glycoproteins (including associations non-replicated in QMDiab), while only half of all SomaLogic proteins are glycoproteins. The Panther classification system (http://pantherdb.org/tools/compareToRefList.jsp) reported a significant enrichment of the GO process "endothelial cell differentiation" (GO:0045446): 5 out of 17 annotated proteins were found (expected were 0.32, p=1.45×10-5). 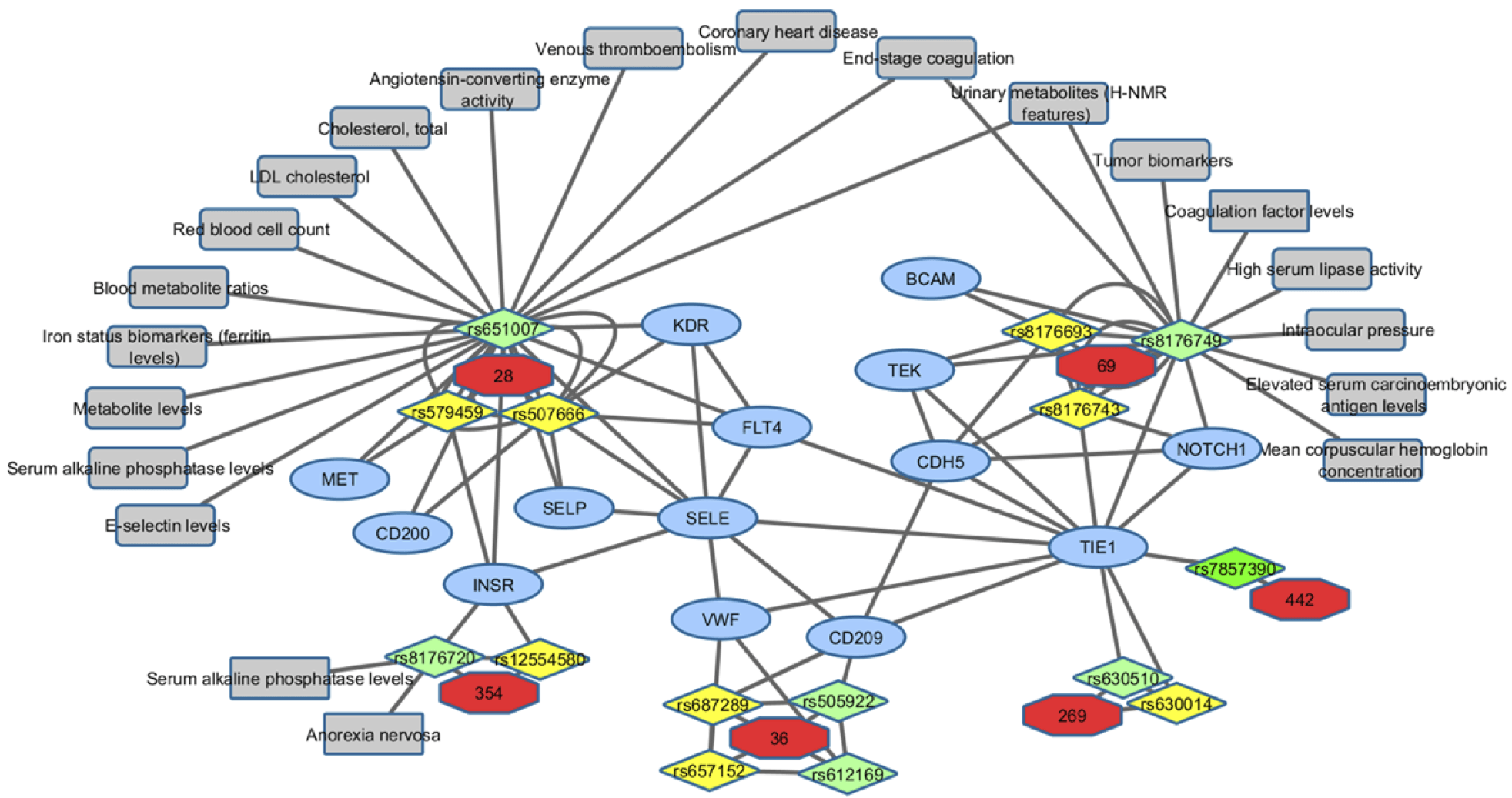
Subnetwork of the ABO locus.Six replicated pQTLs at the ABO gene locus (red), including SNP identifiers that are linked to the the six loci (light green KORA SNPs, yellow QMDiab SNPs, bright green both), pQTLS (blue), proteins with significant partial correlation are connected, associations with GWAS endpoints are shown (grey) [NOTE: the anorexia is a false positive, checked this with the consortium].
The information gathered here is a result of an attempt to keep track of all interesting information that we encountered while investigating these loci. Please bear in mind that the annotation given here is neither complete nor free of errors, and that all information provided here should be confirmed by additional literature research before being used as a basis for firm conclusions or further experiments.






















